Gene Expression Regulation
Cells must regulate which parts of their DNA is being read and used to create proteins. Prokaryotes contain operons, which are segments of DNA that control which segments of DNA will be used to create proteins.
Operons contain:
- The promoter region is the part of DNA that RNA polymerase binds to and begins transcription.
- The operator region controls whether RNA polymerase binds to the promoter. A regulatory protein binds to the operator region.
- Structural genes are the sequences of DNA that code for the proteins being created.
- The regulatory gene synthesizes a regulatory protein, which binds to the operator region. Based on whether the regulatory protein is present or not, the DNA will either be transcribed or inhibited from doing so. There are two types of regulatory proteins:
- Repressor proteins block RNA polymerase from attaching to the promoter. This leads to negative regulation because transcription can only begin when the repressor protein is not present.
- Activator proteins make it easier/promote the binding of RNA polymerase to the promoter. This leads to positive regulation because transcription can easily occur when they are present.
In bacteria, there are three common examples of gene regulation.
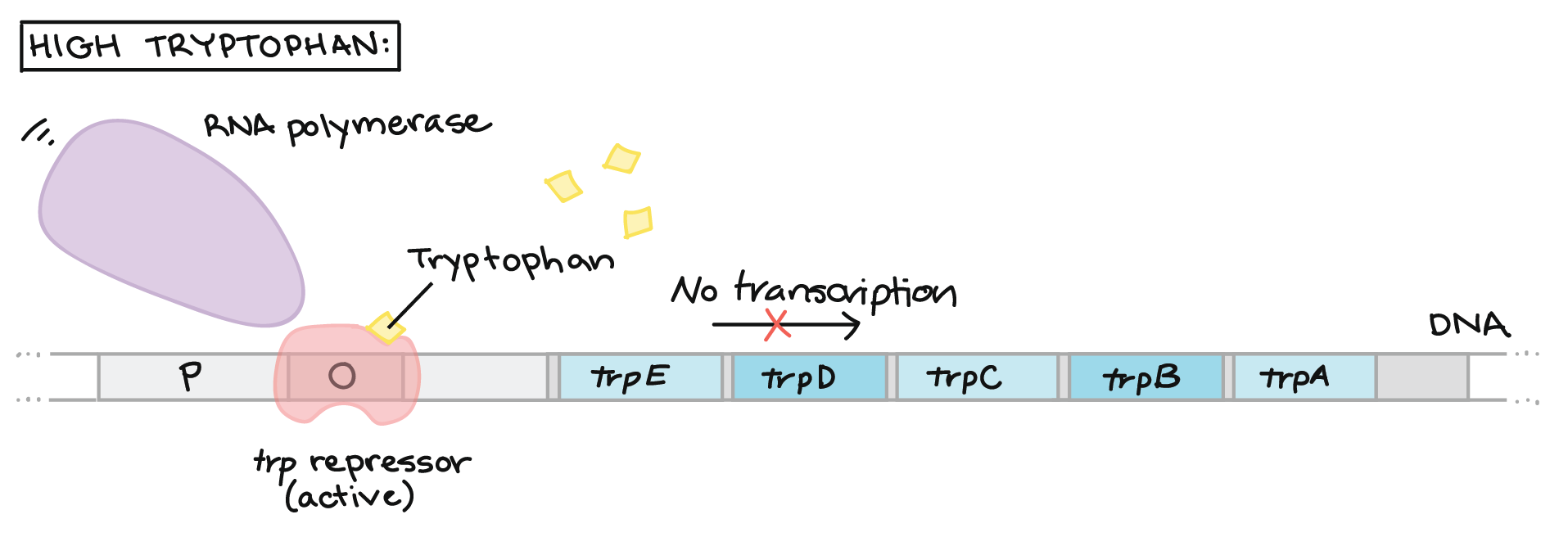
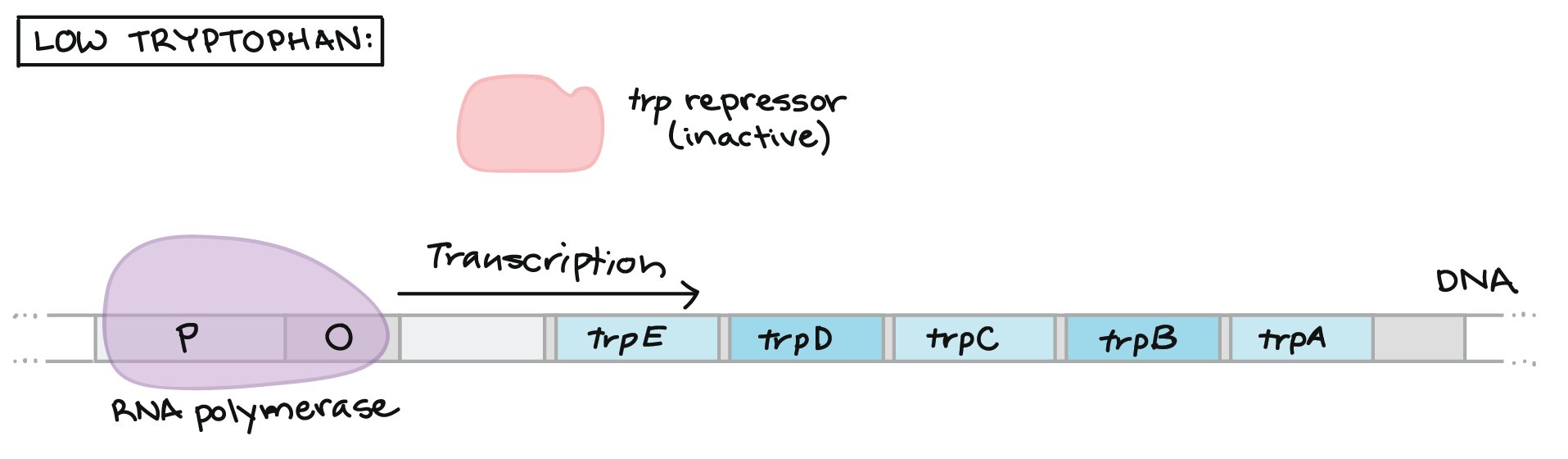
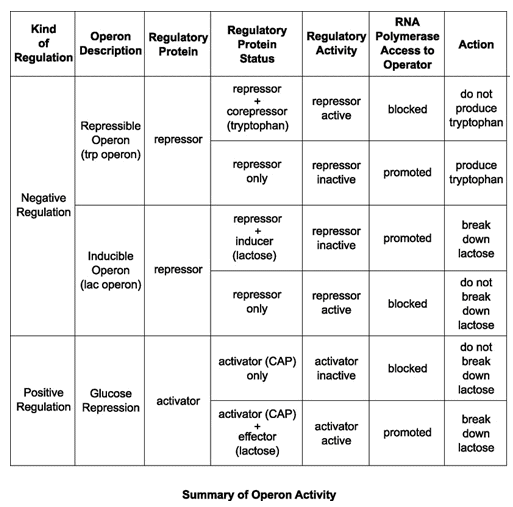
The first is the trp operon. The trp operon codes for enzymes necessary for the synthesis of tryptophan, which is an amino acid. When the bacteria can readily find and absorb tryptophan from their environment, the bacteria does not need to waste energy by trying to produce its own. Some of the tryptophan binds to the trp repressor, causing it to become active and bind to the operator region of the operon. Tryptophan is acting as a corepressor in this case. Since the trp repressor has binded to the operator region, RNA polymerase will not bind to the promoter and the structural genes will not be used to create proteins. The enzymes that the structural genes encode for are called repressible enzymes and the operon is called a repressible operon.
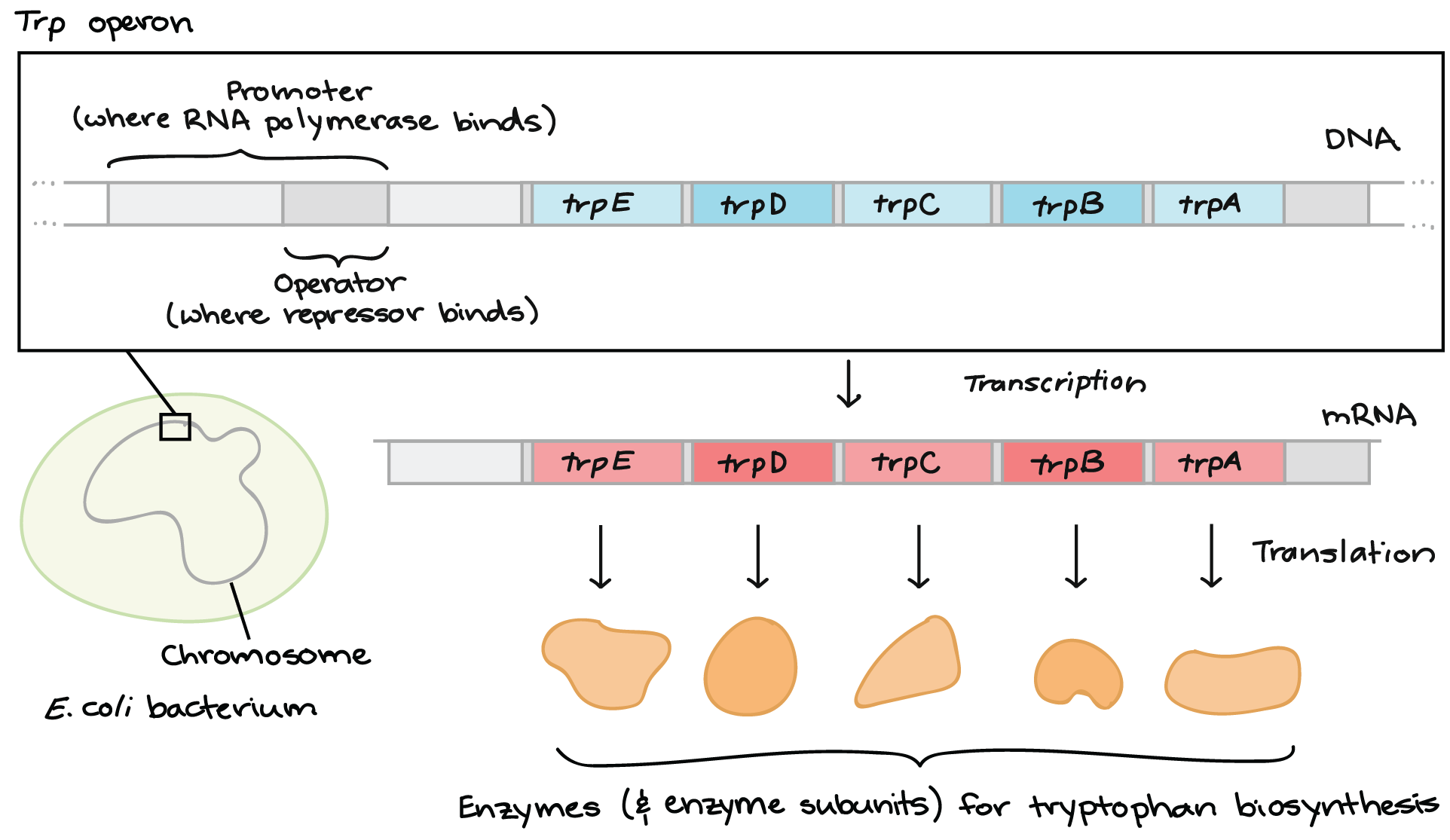
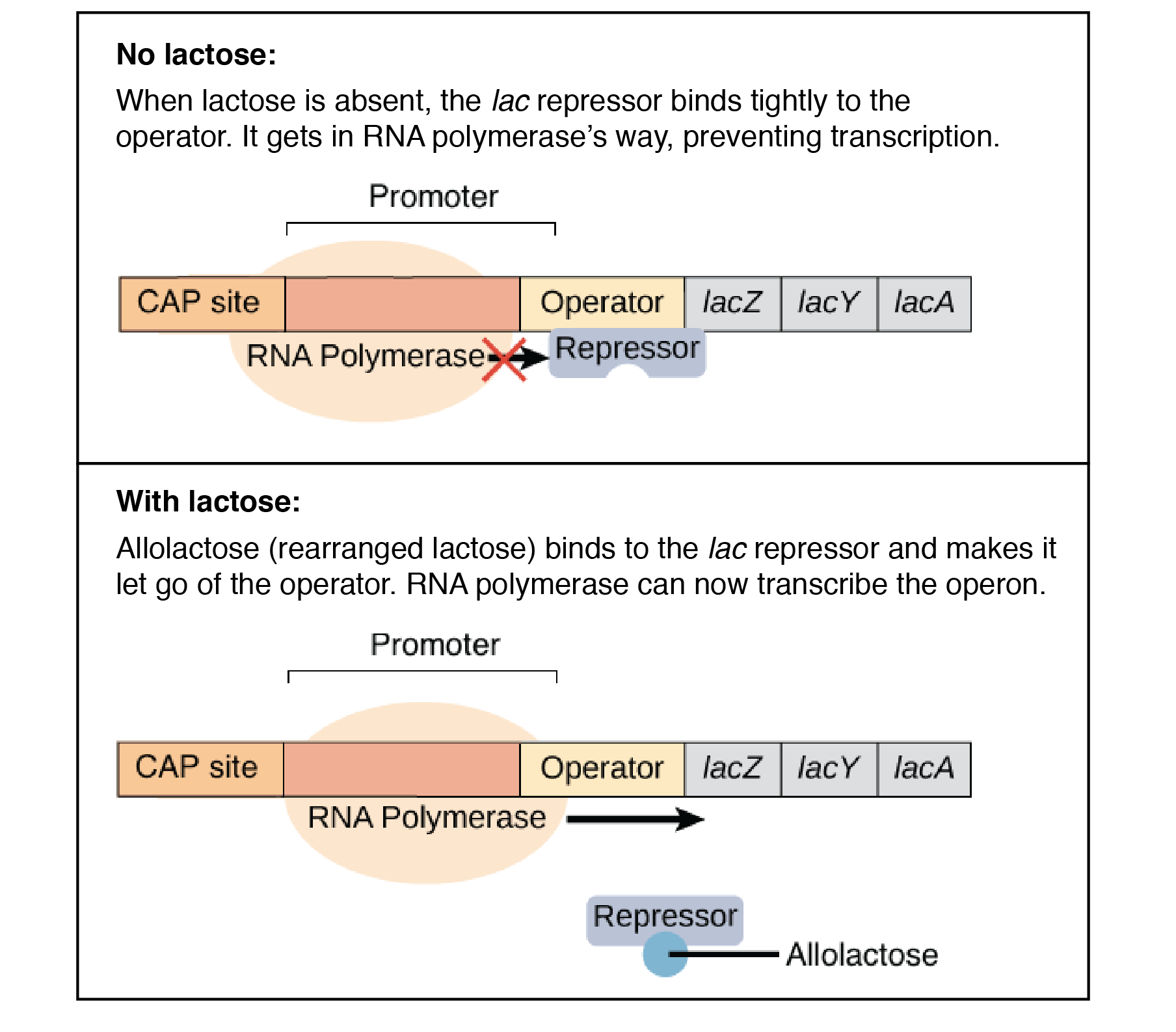
The second example is the lac operon, which is, in a way, the opposite of the trp operon. The lac operon codes of enzymes necessary for the uptake and breakdown of lactose. Unlike trp, the cell will only want to expend energy to breakdown lactose when lactose is available. The regulatory gene in the operon synthesizes an active repressor protein that immediately binds to the operator. When there is little to no lactose present, the RNA polymerase will be unable to bind to the promoter and the structural genes will not be used to produce the lactose digesting enzymes. However, when lactose is available, some of the lactose binds to the repressor protein and the repressor is inactivated. This allows RNA polymerase to bind to the promoter region and synthesize the enzymes. The enzymes that the structural genes encode for are called inducible enzymes and the operon is called an inducible operon.
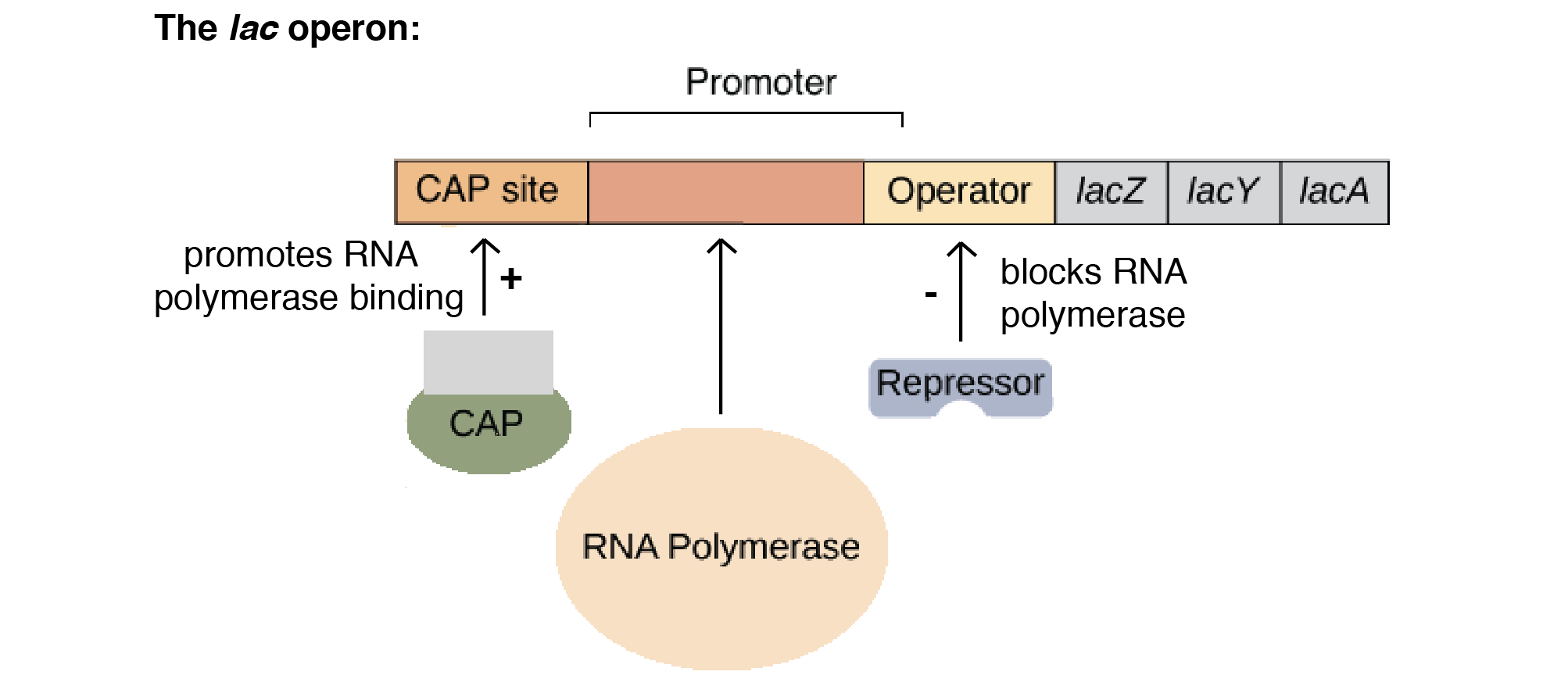
Glucose repression plays a part in the lac operon. When both glucose and lactose are available, the cell prefers glucose as an energy source, and must prevent lactose from being digested. When only lactose is present, however, the cell must allow the lactose digesting enzymes to be synthesized. To make this possible, the cell uses an activator, which is a regulatory protein that binds to the operator and promotes RNA polymerase binding when glucose is available. This is an example of positive regulation.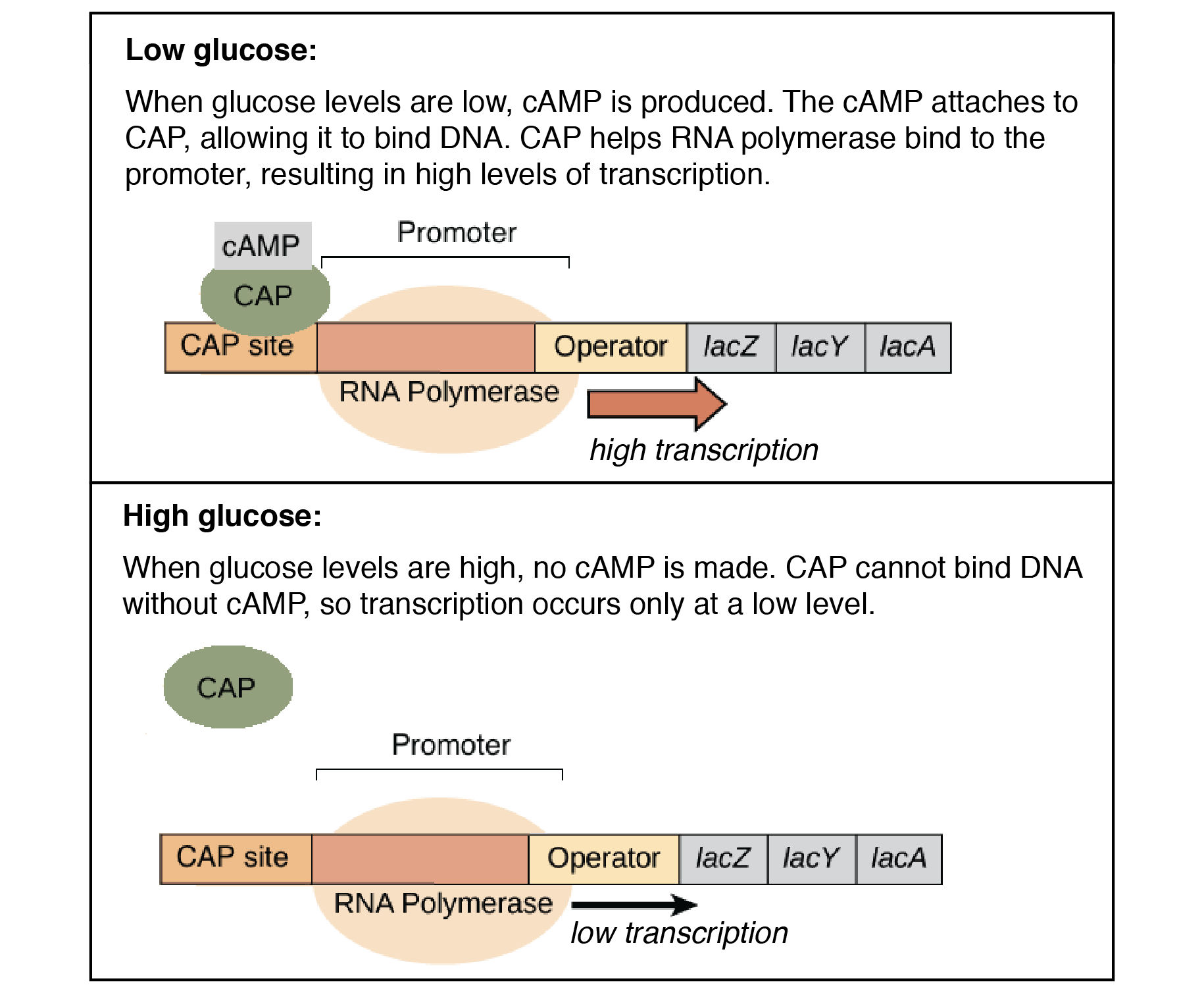
Unlike prokaryotes, eukaryotes are much more complex and require much more complicated methods of gene regulation. Some of the mechanisms for DNA regulation in eukaryotes are listed below:
- DNA methylation is when methyl (CH3) groups attach to the DNA. This makes it more difficult for the transcription factors to access the DNA and can cause long-term inactivation of genes.
- Histone modification is when there are changes in the histone protein organization in the DNA double helix. In DNA acetylation, the histones loosen up and transcription is activated. In DNA methylation, the histones tighten and transcription is inactivated.
- X inactivation only occurs in female mammals. In each cell of female embryos, one of the two X sex chromosomes is randomly inactivated. Any descendants of the cell will contain the same inactivated X chromosome.
